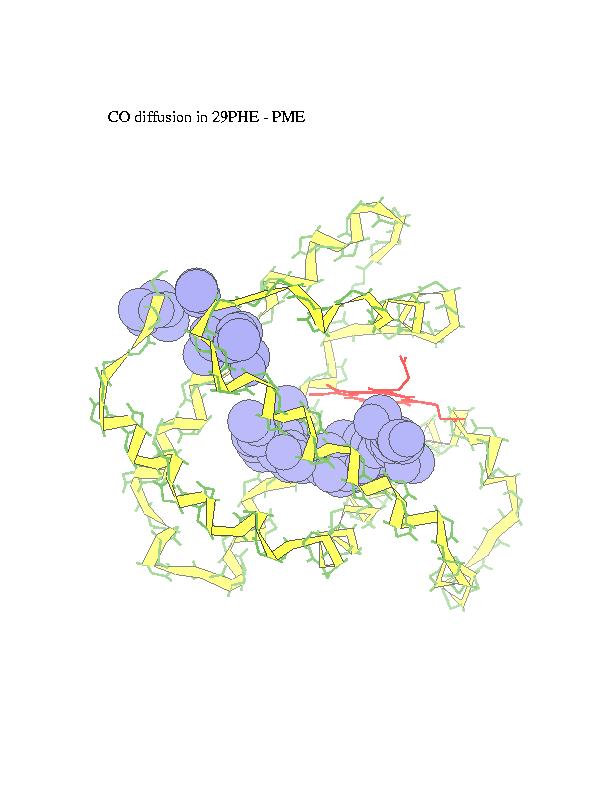
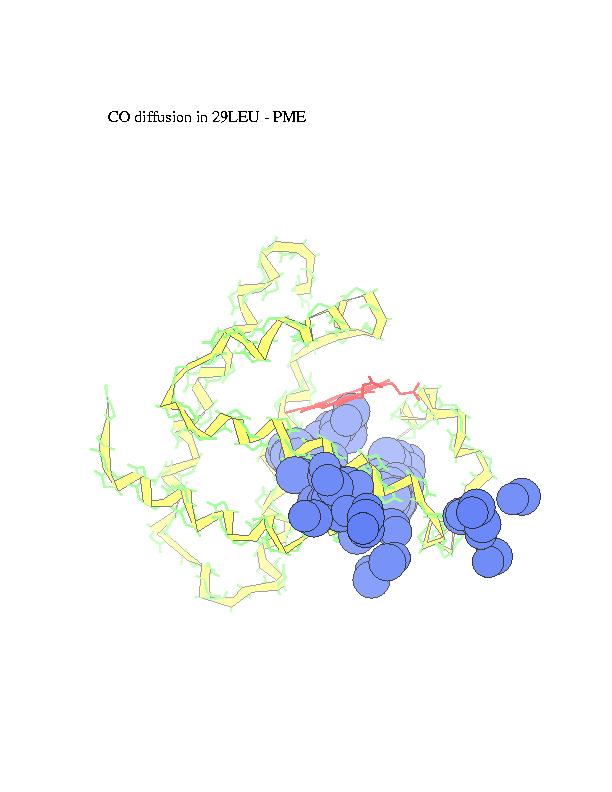
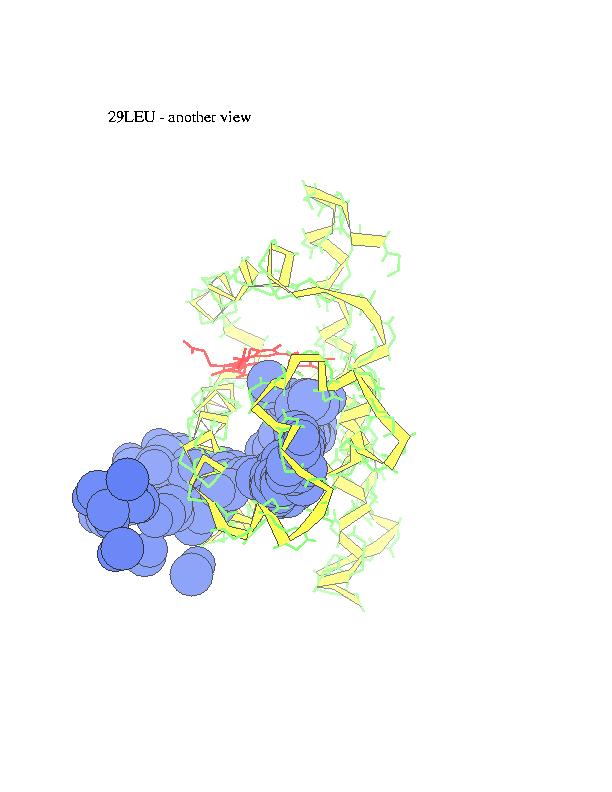
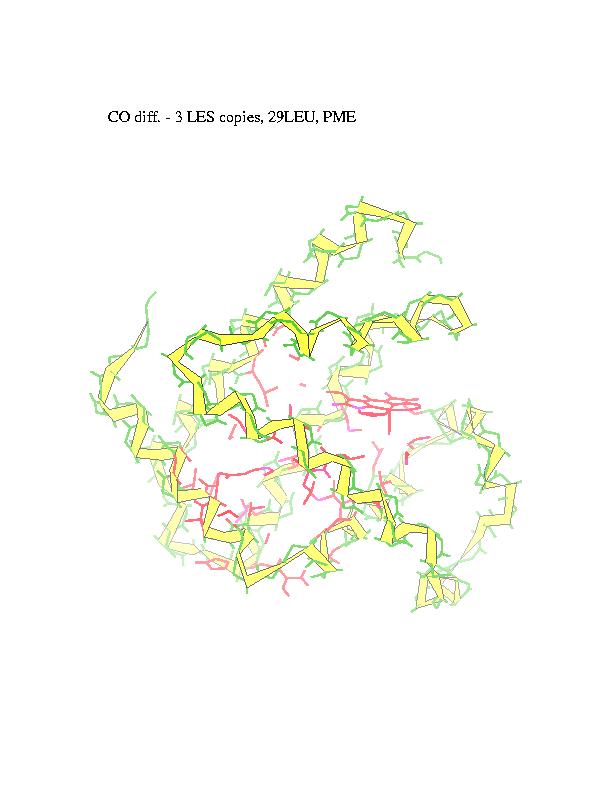
Ligand diffusion pathways through myoglobin and its mutant (Phe29), as observed in MD simulations performed using MOIL package . Locally Enhanced Sampling method by R. Elber, which employs multiple ligand copies, or standard MD with a single ligand in a 'shaky' protein environment are used to enable ligand escape in the sub-nanosecond trajectories.
The subsequent positions of carbonmonoxy ligand with respect to the
protein, as ligand escapes from the heme (marked in red) to the external
medium, are recorded and overlapped in order to obtain a suggestive view
of the trajectory. CO is represented in the figures by blue spheres. In
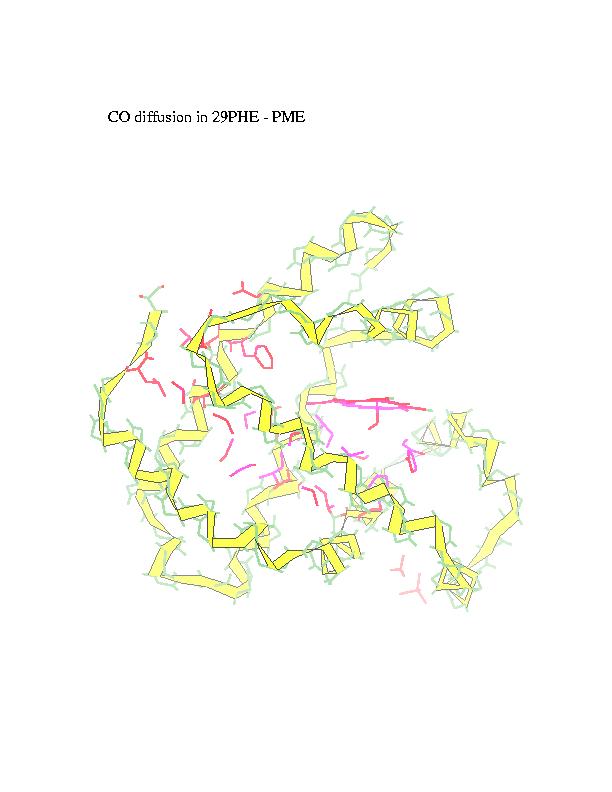
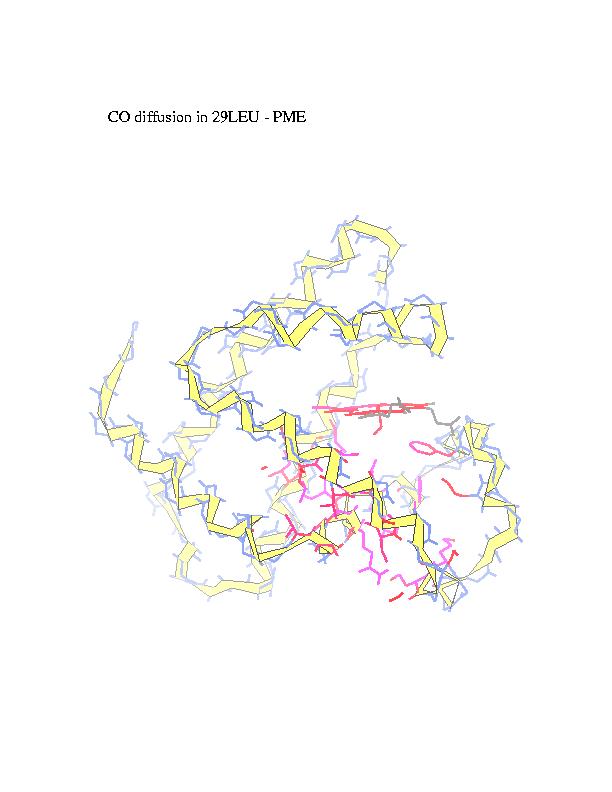
alternative representation of the CO pathway all the residues in contact
with carbon monoxide are marked in red (intensity of the color represents
number of collisions).
The protein threading problem is the assignment of a sequence to a fold (3D structure), according to an energy (scoring function) value. Threading is very useful in protein recognition, especially when searches based on sequence similarity are unable to identify related proteins that are functionally and structurally characterized.
Most commonly, a reduced representation of protein structure (e.g. point
per amino acid residue) and an inter-residue contact potentials are employed
in threading algorithms. Such potential energy (scoring) functions may
be derived by variety of means, for example from the observed frequencies
of contacts. An alternative approach employs Linear Programming to obtain
an energy function recognizing exactly all the proteins included in the
training set.
One of the key difficulties in the threading problem is the use and design of appropriate energy functions, that recognize the correct or similar fold and do it efficiently. The threading problem is known to be NP hard, when standard pairwise potentials are used.
We introduced (joint work with R. Elber) a new class of `contact shell' potentials with energy that depends only on the identity of the site residue and the number of its neighbors in the first and second contact shell. We call it THOM (THreading Onion Model). The model is local, and thus enables efficient threading search, and it is demonstrated to have recognition capacity comparable to that of pairwise potentials. THOM model has been implemented in our package for protein recognition, called LOOPP.