Please click the links to view the results:
Secondary Structure:
For the domains in RPA whose tertiary structure was available, the
secondary structure and solvent accessibility was visualized using the
POLYVIEW server.
Otherwise, the secondary structure and solvent accessibility of amino acid
residues were predicted using our
SABLE protein
structure prediction server. POLYVIEW was again used to combine the results.
Please click the links to view the results: ![]() RPA14
,
RPA14
, ![]() RPA32
,
RPA32
, ![]() RPA70.
RPA70.
Tertiary structure:
The different entries in the Protein Data Bank are listed below.
1. 1L1O
: RPA trimerization core containing RPA70 C-terminal domain, RPA32 central domain and RPA14.
2. 1JMC
: RPA70 DNA binding domain bound to a DNA single-strand.
3. 1FGU
: RPA70 Central domain.
4. 1QUQ
: RPA32 Central domain and RPA14.
5. 1DPU
: RPA32 C-terminal domain bound to Uracil DNA Glycosylase (UNG2).
Note: For RPA14: Full tertiary structure (1L1O,1QUQ) has been determined.
For RPA32: N-terminal structure is not clear. Central domain (1L1O,1QUQ) and C-terminal (1DPU) are available.
For RPA70: N-terminal structure is not clear. Central domain (1JMC,1FGU) and C-terminal (1L1O) are available.
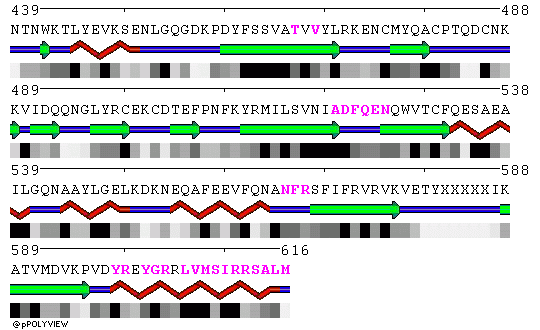
POLYVIEW representation of the trimeric core crystal structure 1L1O are included below. Residues in contact in the trimer are highlighted in magenta in chains A, B and C, respectively.
Chain A:

Chain B:
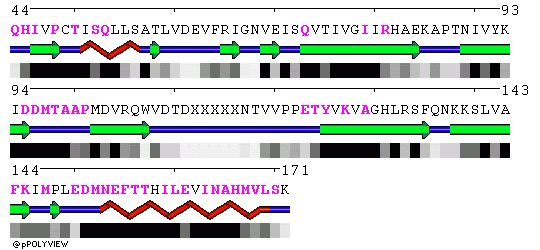
Chain C: